From PDB to Pose: Integration of Nanome's MARA and Cresset's Flare
Every small‑molecule project starts with a question: can we place the right chemistry in the right pocket, quickly and without giving up scientific control? This demonstration follows that exact journey. Fujitsu, our reseller and systems integrator in Japan, set out to connect MARA with Flare™ from Cresset®, to enable scientists to move from an accession code to validated poses in a single conversation. The images below are frames from a Fujitsu recording of that workflow in action.
Why Flare
Flare is Cresset's complete CADD solution for ligand-based and structure-based design, high-resolution 3D visualization and in-depth analysis of ligand series and biological targets. Combining robust computational methods with AI/ML, it is widely used across pharma, agrochem and biotech companies to generate novel, active molecules with optimum efficiency. Scientists choose Flare because it balances prediction quality, interactive control and traceability with ease of use and cost efficiency. In this integration, especially two tools Pyflare and the Flare Python API helped Fujitsu a lot. The Python API and the pyflare module facilitates simple, third-party integration, and the advanced scope of methods including docking and scoring, Electrostatic Complementarity™, Free Energy Perturbation enable scientists to make fast, informed decisions. Parameters are explicit, results are inspectable, and workflows are proven in production.
Why this milestone matters
MARA already orchestrates open source tools such as RDKit, P2Rank, and AutoDock Vina, which is ideal for rapid prototyping. This integration shows the same agentic approach working cleanly with a closed source, enterprise grade platform. That unlocks governed data paths, vendor support, and the compliance posture that global pharma and biotech expect. In short, MARA can run commercial engines side by side with open source utilities inside one private, audited pipeline.
Protein context: FKBP12 and 1FKG
1FKG is human FKBP12, the FK506 binding protein that acts as a peptidyl prolyl cis/trans isomerase and is present in many tissues. FKBP12 forms complexes with tacrolimus and sirolimus, which modulate calcineurin and mTOR pathways. This makes it relevant to immunology, transplant medicine, and oncology research. The 1FKG entry contains a high affinity synthetic ligand, SB3, and was solved at 2.0 Å resolution. It is a practical demo target because the pocket is well defined, the protein is small and stable, and there is strong benchmarking data for docking and SAR. Using 1FKG lets us show how MARA moves a real structure through preparation, cavity detection, and docking, then produces poses that scientists can inspect or compare with the crystallographic reference.
How the integration works at a glance
MARA orchestrates each action, passes correctly formatted files into Flare, and records every step with inputs and outputs. This hand-off is performed programmatically via pyflare and the Flare Python API, allowing automated execution of protein preparation, docking, and scoring without brittle manual steps. Scientists keep control of parameters while skipping brittle file handoffs. The flow runs on premises or in a private cloud with SSO and audit logs.
Walkthrough of the 1FKG demo
1) Retrieve the structure
A user asks MARA to download PDB 1FKG. MARA pulls the entry and stores it as 1fkg.pdb, then presents an interactive 3D preview so the user can confirm the target before proceeding.
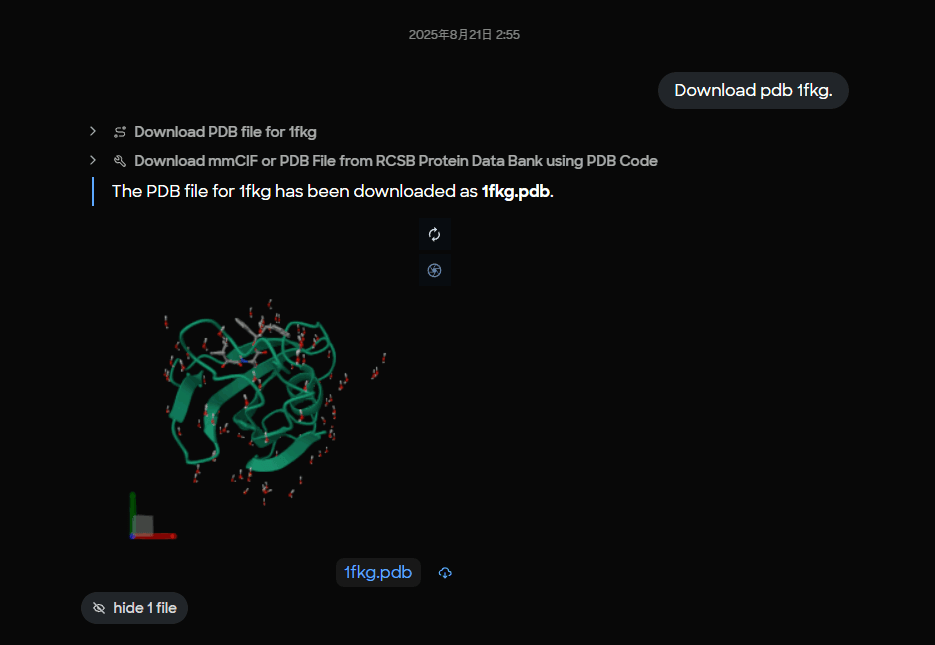 Figure 1. MARA interface showing successful download of PDB 1FKG with 3D molecular visualization preview of the FKBP12 protein structure.
Figure 1. MARA interface showing successful download of PDB 1FKG with 3D molecular visualization preview of the FKBP12 protein structure.
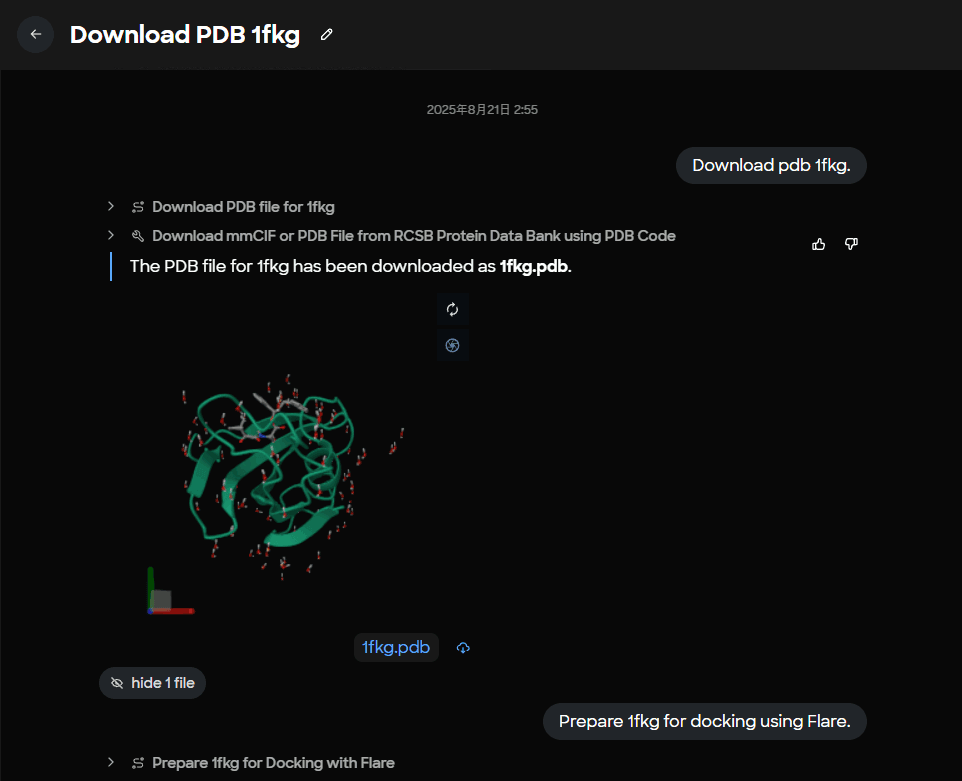 Figure 2. Task completion panel displaying the saved 1fkg.pdb file with confirmation message for PDB structure retrieval.
Figure 2. Task completion panel displaying the saved 1fkg.pdb file with confirmation message for PDB structure retrieval.
2) Prepare the protein with Flare
MARA runs a Flare protein preparation step on 1fkg.pdb, producing a docking‑ready file named 1fkg_P.pdb. Preparation typically covers hydrogen addition, protonation assignment, residue and ligand filtering, and other conditioning that makes a structure suitable for docking.
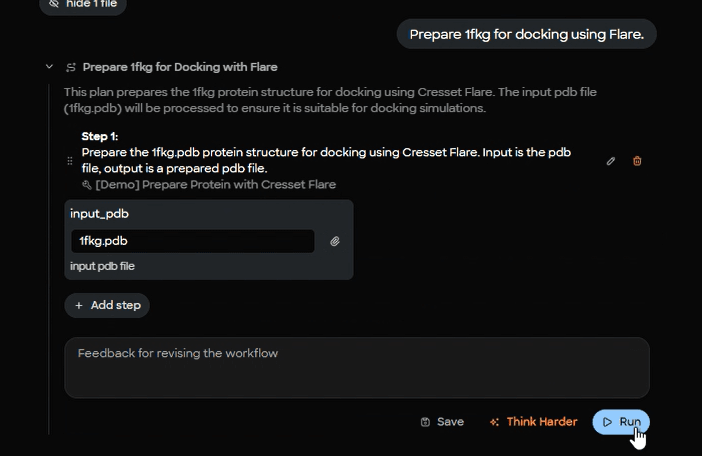 Figure 3. MARA plan editor interface for Flare protein preparation workflow with input parameter specification for 1fkg.pdb.
Figure 3. MARA plan editor interface for Flare protein preparation workflow with input parameter specification for 1fkg.pdb.
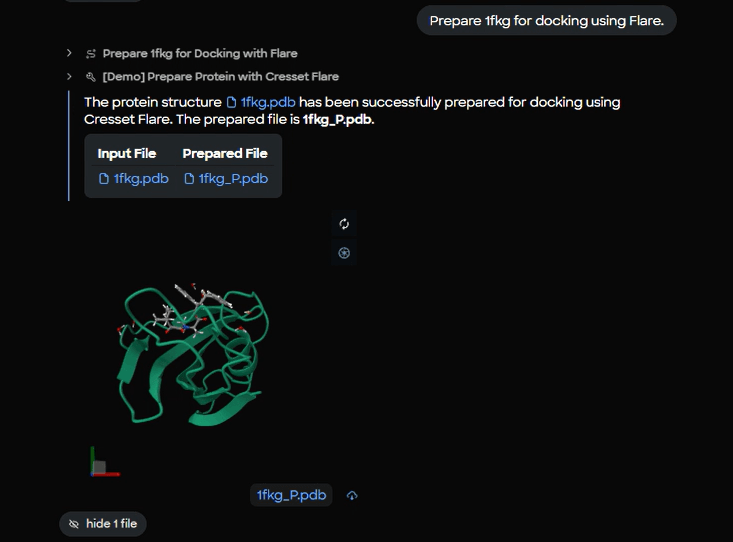 Figure 4. Protein preparation completion panel showing input file (1fkg.pdb) and prepared output file (1fkg_P.pdb) with live 3D molecular preview.
Figure 4. Protein preparation completion panel showing input file (1fkg.pdb) and prepared output file (1fkg_P.pdb) with live 3D molecular preview.
3) Detect likely binding pockets
To guide search space selection, MARA calls a pocket predictor on the prepared protein. In this demo Fujitsu used P2Rank. The result includes a probability and a pocket center.
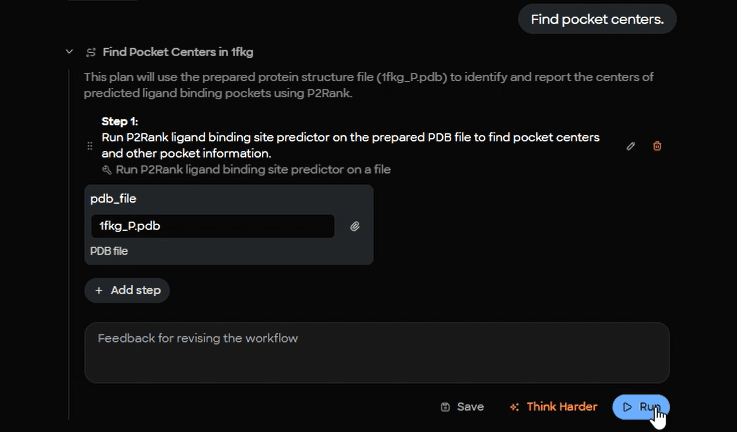 Figure 5. P2Rank cavity detection setup interface showing input parameters for binding pocket prediction on prepared 1fkg_P.pdb structure.
Figure 5. P2Rank cavity detection setup interface showing input parameters for binding pocket prediction on prepared 1fkg_P.pdb structure.
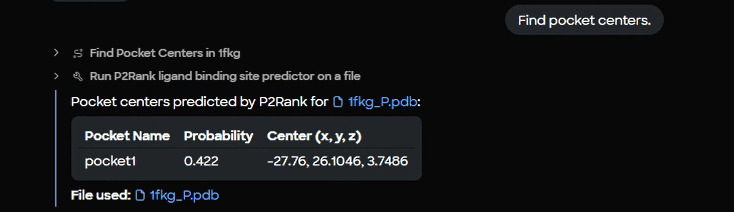 Figure 6. P2Rank results table displaying identified binding pocket with probability score (0.422) and 3D center coordinates (-27.76, 26.1046, 3.7486).
Figure 6. P2Rank results table displaying identified binding pocket with probability score (0.422) and 3D center coordinates (-27.76, 26.1046, 3.7486).
4) Identify any co‑crystallized ligands
MARA parses the macromolecule to list three‑letter ligand codes. For 1FKG, the code SB3 is detected.
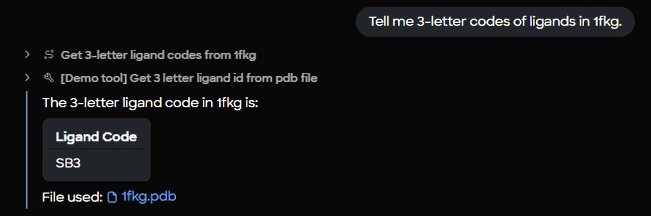 Figure 7. Ligand extraction interface and results panel confirming detection of co-crystallized ligand SB3 from the 1fkg.pdb structure.
Figure 7. Ligand extraction interface and results panel confirming detection of co-crystallized ligand SB3 from the 1fkg.pdb structure.
5) Retrieve the reference ligand as SDF
With the code in hand, MARA downloads the SDF for SB3 for use as a docking reference and displays a 3D preview.
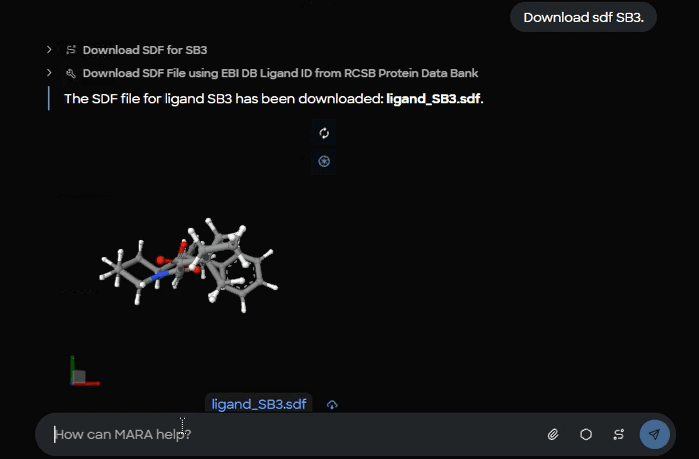 Figure 8. Ligand download confirmation showing saved ligand_SB3.sdf file with 3D structural preview of the SB3 reference compound.
Figure 8. Ligand download confirmation showing saved ligand_SB3.sdf file with 3D structural preview of the SB3 reference compound.
6) Dock SB3 into 1FKG using Flare
MARA passes the prepared protein, the ligand SDF, and the cavity center into Flare with explicit box dimensions and pose count. The demo shows these parameters: center x = -27.76, y = 26.1046, z = 3.7486, box length 16 Å, box width 16 Å, box height 16 Å, maximum conformations 10.
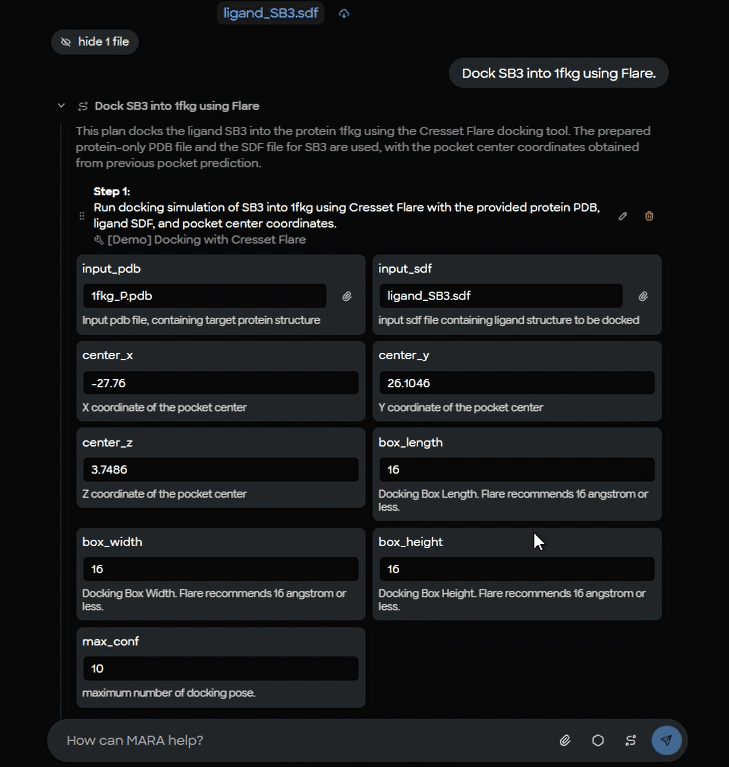 Figure 9. Flare docking parameter setup form showing input files (1fkg_P.pdb protein, ligand_SB3.sdf), binding site coordinates, and docking box dimensions (16 ų).
Figure 9. Flare docking parameter setup form showing input files (1fkg_P.pdb protein, ligand_SB3.sdf), binding site coordinates, and docking box dimensions (16 ų).
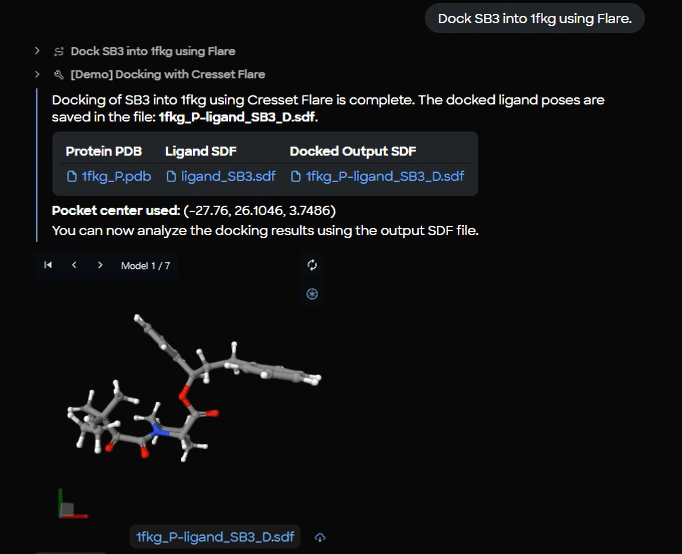 Figure 10. Docking completion results showing successful SB3 docking into 1fkg with output file (1fkg_P_ligand_SB3_D.sdf) and 3D viewer displaying Model 1 of 7 generated poses.
Figure 10. Docking completion results showing successful SB3 docking into 1fkg with output file (1fkg_P_ligand_SB3_D.sdf) and 3D viewer displaying Model 1 of 7 generated poses.
7) Create a combined complex for review
For easier sharing and visual inspection, MARA merges the prepared protein with all docked poses into a single multi‑frame PDB.
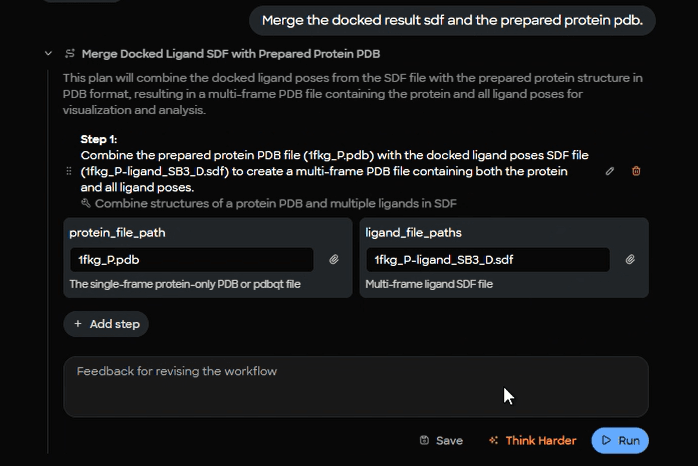 Figure 11. Complex merging workflow interface showing combination of prepared protein file (1fkg_P.pdb) with docked ligand poses (1fkg_P_ligand_SB3_D.sdf).
Figure 11. Complex merging workflow interface showing combination of prepared protein file (1fkg_P.pdb) with docked ligand poses (1fkg_P_ligand_SB3_D.sdf).
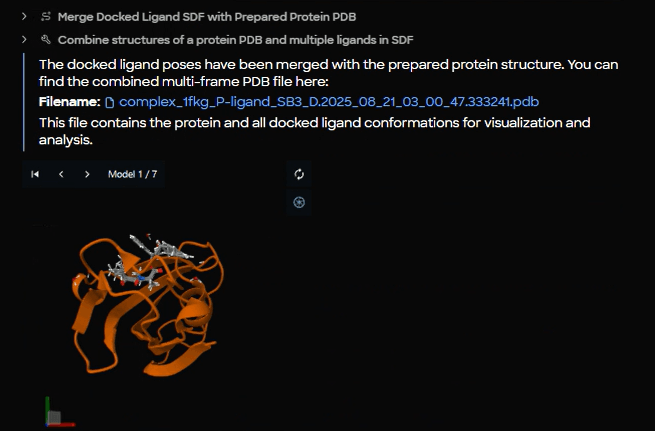 Figure 12. Merged complex output panel displaying the combined PDB file (complex_1fkg_P_ligand_SB3_D.2025_08_21_03_00_47.333241.pdb) with orange cartoon protein structure and SB3 poses.
Figure 12. Merged complex output panel displaying the combined PDB file (complex_1fkg_P_ligand_SB3_D.2025_08_21_03_00_47.333241.pdb) with orange cartoon protein structure and SB3 poses.
8) Browse through poses
The complex opens with an interactive model selector so the scientist can step through poses and zoom into the pocket.
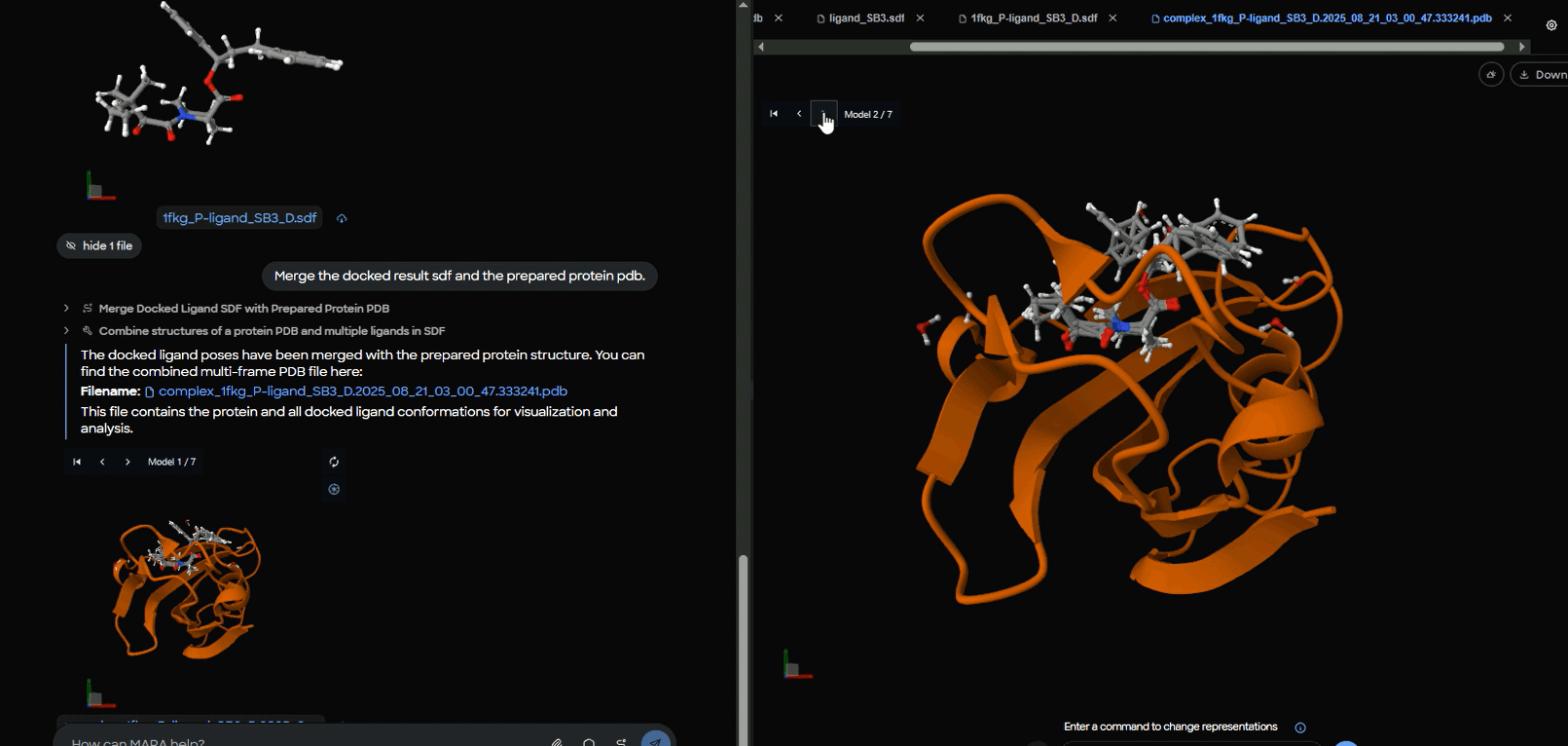 Figure 13. Interactive pose browser showing protein-ligand complex with model selector set to "Model 2 of 7" for systematic evaluation of different docking conformations.
Figure 13. Interactive pose browser showing protein-ligand complex with model selector set to "Model 2 of 7" for systematic evaluation of different docking conformations.
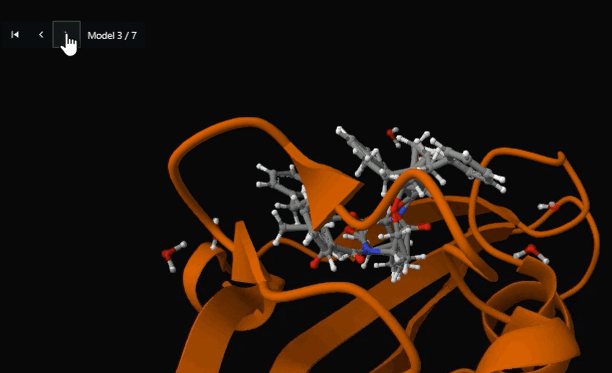 Figure 14. Detailed binding site view showing SB3 ligand positioned within the FKBP12 active site cavity (Model 3 of 7) for structure-activity relationship analysis.
Figure 14. Detailed binding site view showing SB3 ligand positioned within the FKBP12 active site cavity (Model 3 of 7) for structure-activity relationship analysis.
Why this matters for teams
This pattern saves time without hiding science. Parameters remain visible, intermediate files are preserved, and the full run is repeatable. Teams can template the sequence and apply it to other targets, then extend it with enumeration, rescoring, MMGBSA or FEP, PLIF analysis, and immersive inspection in Nanome. The integration runs in a controlled environment so data never leaves your boundary.
Acknowledgments and next steps
Thank you to Fujitsu for the implementation and demo, and to Cresset for building Flare, a powerful ligand and structure‑based design platform. Flare is a trademark of Cresset. All other names are the property of their respective owners.
Get in touch
If you are a Japanese biopharma company and would like to explore this workflow, please reach out directly to Fujitsu. For organizations in the US or Europe interested in seeing MARA and Flare in action, contact us to arrange a demonstration.