Nanome 2.0: A Deep Dive into the Technical Foundations
Since launching Nanome AI (along with with Nanome 2.0) in early access, we’ve explored Nanome 2.0’s new scene menu and the enhanced permissioning system in previous blog posts. Today, we’re excited to take you on a deep dive into the technical foundations that make Nanome 2.0 a significant evolution from our 1.x versions.
Built from the ground up, Nanome 2.0 embodies the many lessons we’ve learned over the years. This isn’t just an update; it’s a complete reimagining of our platform’s core architecture to provide a more powerful, efficient, and collaborative experience. Let’s delve into the technical innovations and foundational changes that set Nanome 2.0 apart.
Reimagined Data Persistence and Seamless Collaboration
One of the most impactful enhancements in Nanome 2.0 is the overhaul of our data persistence and sharing mechanisms. In the 1.x versions, users often had to manually save workspaces, track who saved what, and manage room sessions—all of which could interrupt the collaborative workflow.
With Nanome 2.0, we’ve introduced cloud-based data persistence. Your molecular data and workspace settings are now automatically saved and synchronized in the cloud, eliminating the need for manual saves or complex workspace management. This advancement enables truly seamless collaboration, allowing teams to focus on innovation rather than logistical hurdles.
Advanced Rendering Engine Optimized for Mobile XR (All-In-One Devices)
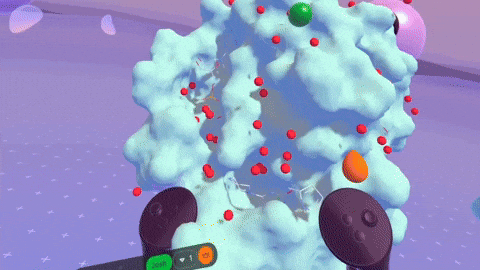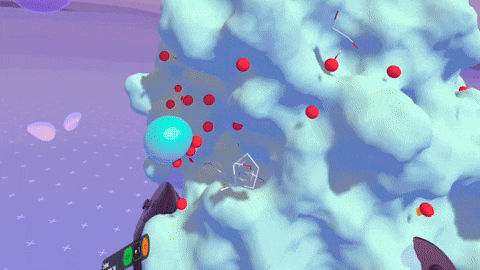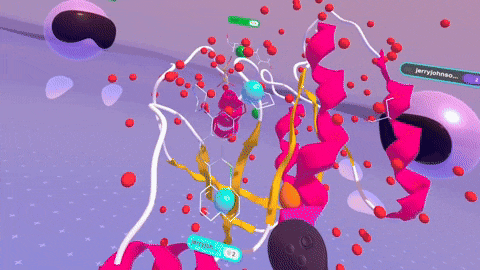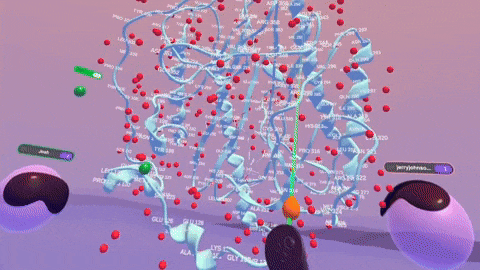
Rendering complex molecular structures on spatial computing platforms demands both performance and visual fidelity. Back in the Nanome 1.x era, our rendering engine was crafted for PC VR, specifically with devices like the HTC Vive and Oculus Rift CV1 in mind. We later stretched its capabilities to embrace mobile platforms. But with Nanome 2.0, we’ve taken a bold leap forward. We’ve rebuilt our rendering engine from the ground up using the Universal Render Pipeline (URP). This game-changing move supercharges Nanome for mobile XR platforms and even unlocks rendering on Apple Silicon devices like the Apple Vision Pro. The result? Smoother, more responsive visuals across a wider array of devices than ever before.
Enhanced Molecular Data Structures for Large-Scale Handling
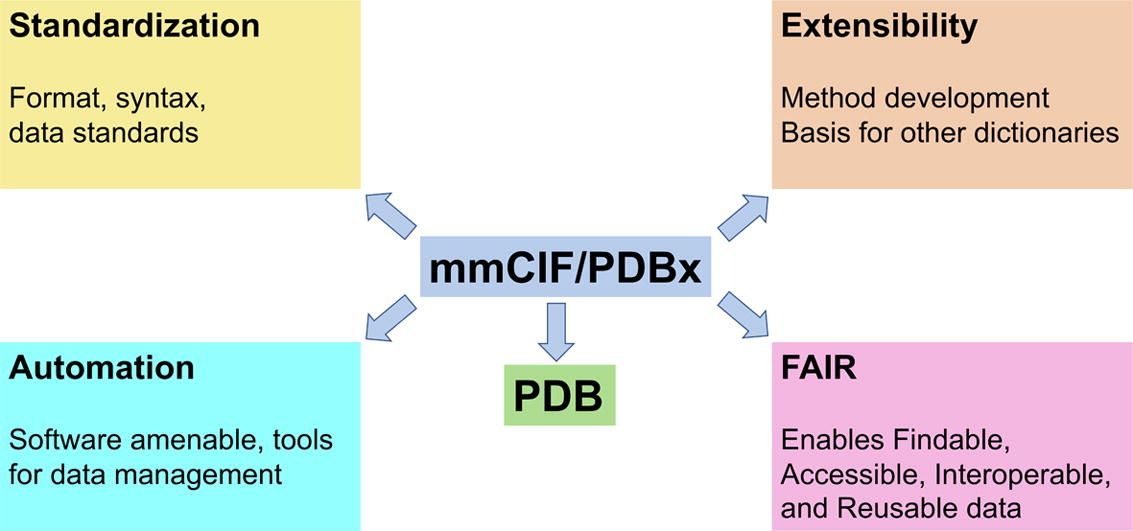
Modern molecular research often involves handling vast and complex datasets. Recognizing this, we’ve made significant strides in optimizing our molecular data structures. Nanome 2.0 introduces a new framework for accessing and mutating molecular data, enabling the platform to efficiently handle much larger atomic files. This improvement is particularly evident in our enhanced support for PDBx/mmCIF standardized files from the RCSB Protein Data Bank. Users can now load and interact with large-scale molecular models more effortlessly, facilitating deeper analysis and exploration without performance compromises.
Source: https://www.sciencedirect.com/science/article/pii/S0022283622001796
Building for the Future While Mindful of Performance
While rebuilding Nanome from the ground up has provided us with a leaner and more efficient platform, we are conscious that adding new features could impact performance over time. Our commitment is to maintain optimal performance as we continue to develop Nanome 2.0, paying close attention at every step to ensure the platform remains both powerful and user-friendly.
We understand that performance is paramount for our users’ workflow and the integrity of their research. Therefore, we are dedicated to ongoing optimization as we expand Nanome’s capabilities.
Optimized for Tomorrow’s Science
Nanome 2.0 represents a bold step forward in our mission to empower scientists, educators, and students with cutting-edge tools for molecular visualization and collaboration. By reengineering our platform with a focus on data persistence, rendering performance, and molecular data handling, we’ve laid a strong foundation for future innovations. We invite you to experience the technical advancements of Nanome 2.0 and join us in shaping the future of molecular science with spatial computing.